5.17. Run DESID CMAQ on hbv3_120 using the beeond filesystem#
5.17.1. Run CMAQ for DESID#
Edit the DESID Namelist#
Edit the CMAQ DESID Chemical Species Control File
cd /shared/build/openmpi_gcc/CMAQ_v54/CCTM/scripts/BLD_CCTM_v54_gcc
cp CMAQ_Control_DESID_cb6r5_ae7_aq.nml CMAQ_Control_DESID_cb6r5_ae7_aq_RED_EGU_POINT_NY.nml
vi CMAQ_Control_DESID_cb6r5_ae7_aq_RED_EGU_POINT_NY.nml
Add the following lines to the bottom of the file according to the DESID Tutorial Instructions
https://github.com/USEPA/CMAQ/blob/main/DOCS/Users_Guide/Tutorials/CMAQ_UG_tutorial_emissions.md#scale_stream (place the line before the / file marker)
! PT_EGU Emissions Scaling reduce PT_EGU emissions in New York by 25%. Note, to reduce the emissions by 25% we use DESID to multiply what had been 100% emissions by .75, so that the resulting emissions is reduced by 25%.
'NY' , 'PT_EGU' ,'All' ,'All' ,'All' ,.75 ,'UNIT','o',
Activate DESID Diagnostics
Create a DESID Control File and edit it to define NY as a region, and activate DESID emissions diagnostics Define NY as a region in the DESID Region Definitions
cp CMAQ_Control_DESID.nml CMAQ_Control_DESID_RED_EGU_POINT_NY.nml
vi CMAQ_Control_DESID_RED_EGU_POINT_NY.nml &
Modify the following section to use the NY region that is specified in the CMAQ_MASKS file, note the CMAQ_MASKS file is defined in the DESID Run script.
&Desid_RegionDef
Desid_Reg_nml =
! Region Label | File_Label | Variable on File
! 'EVERYWHERE' ,'N/A' ,'N/A',
'NY' ,'CMAQ_MASKS' ,'NY',
!<Example> 'ALL' ,'ISAM_REGIONS','ALL',
/
Create two stream family definitions, one that includes all point source emissions, and the second that only contains PT_EGU
!------------------------------------------------------------------------------!
! Emissions Scaling Family Definitions !
! This component includes definitions for families of emission streams and !
! region combinations. !
!------------------------------------------------------------------------------!
&Desid_StreamFamVars
Desid_N_Stream_Fams = 2 ! Exact number of stream family definitions
Desid_Max_Stream_Fam_Members = 20 ! Larger than the number of streams on all
! family definitions
/
&Desid_StreamFam
! For emission streams available in several run scripts under CCTM/scripts
StreamFamilyName(1) = 'PT_SOURCES'
StreamFamilyMembers(1,1:8)= 'PT_NONEGU','PT_OTHER', 'PT_AGFIRES', 'PT_FIRES', 'PT_RXFIRES', 'PT_OTHFIRES', 'PT_OILGAS','PT_CMV_C1C2'
StreamFamilyName(2) = 'PT_EGUS'
StreamFamilyMembers(2,1:1)= 'PT_EGU'
activate DESID diagnostics to report the reduction in PT_EGU emissions.
Note, if you define only one diagnostic rule, you must comment out all other rules.
&Desid_DiagVars
Desid_N_Diag_Rules = 1 ! Exact Number of Diagnostic Rules Below
Desid_Max_Diag_Streams=20 ! Maximum number of species variables on all rules
! below (do not count expansions)
Desid_Max_Diag_Spec = 80 ! Maximum number of species variables on all rules
! below (do not count expansions)
/
! Create a diagnostic of the sum of the components of the PT_SOURCES
! family (defined in the stream family section). This file will be column sums
! and will include all the emitted species as long as they appear on at least
! one of the streams within PT_SOURCES.
Desid_Diag_Streams_Nml(1,:)= 'PT_EGUS'
Desid_Diag_Fmt_Nml(1) = 'COLSUM'
Desid_Diag_Spec_Nml(1,:) = 'ALL'
Verify that the settings are correct by comparing to the version in the github repo directory
diff CMAQ_Control_DESID_RED_EGU_POINT_NY.nml /shared/pcluster-cmaq/qa_scripts/workshop/CMAQ_Control_DESID_RED_EGU_POINT_NY.nml
Edit runscript to use DESID Namelist#
Copy the Run script and edit it to use the DESID namelist files
cd /shared/build/openmpi_gcc/CMAQ_v54/CCTM/scripts/
cp run_cctm_2018_12US1_v54_cb6r5_ae6.20171222.2x96.ncclassic.csh run_cctm_2018_12US1_v54_cb6r5_ae6.20171222.2x96.ncclassic_DESID_RED_NY.csh
Change APPL to a new name
vi run_cctm_2018_12US1_v54_cb6r5_ae6.20171222.2x96.ncclassic_DESID_RED_NY.csh
Change APPL
set APPL = 2018_12US1_DESID_REDUCE #> Application Name (e.g. Gridname)
Verify the following emission stream names match the names used in the DESID namelist.
grep STK_EMIS_LAB_00 run_cctm_2018_12US1_v54_cb6r5_ae6.20171222.2x96.ncclassic_DESID_RED_NY.csh
Output
setenv STK_EMIS_LAB_001 PT_NONEGU
setenv STK_EMIS_LAB_002 PT_EGU
setenv STK_EMIS_LAB_003 PT_OTHER
setenv STK_EMIS_LAB_004 PT_AGFIRES
setenv STK_EMIS_LAB_005 PT_FIRES
setenv STK_EMIS_LAB_006 PT_RXFIRES
setenv STK_EMIS_LAB_007 PT_OTHFIRES
setenv STK_EMIS_LAB_008 PT_OILGAS
setenv STK_EMIS_LAB_009 PT_CMV_C1C2
Compare the above settings to those used in the Emission Stream Family defined in the DESID Namelist.
grep -A 2 -B 2 StreamFamilyMembers ./BLD_CCTM_v54_gcc/CMAQ_Control_DESID_RED_EGU_POINT_NY.nml
Output
StreamFamilyName(1) = 'PT_SOURCES'
StreamFamilyMembers(1,1:4)= 'PT_NONEGU','PT_OTHER', 'PT_AGFIRES', 'PT_FIRES', 'PT_RXFIRES', 'PT_OTHFIRES', 'PT_OILGAS','PT_CMV_C1C2'
StreamFamilyName(2) = 'PT_EGUS'
StreamFamilyMembers(2,1:1)= 'PT_EGU'
Note
CMAQ won’t crash if the stream name in CMAQ_Control_DESID_
Update the DESID namelist file names in the run script to use the Reduced PT_EGU and diagnostic instructions.
cd /shared/build/openmpi_gcc/CMAQ_v54/CCTM/scripts
vi run_cctm_2018_12US1_v54_cb6r5_ae6.20171222.2x96.ncclassic_DESID_RED_NY.csh
Modify the namelist setting to use the DESID namelist:
setenv DESID_CTRL_NML ${BLD}/CMAQ_Control_DESID_RED_EGU_POINT_NY.nml
setenv DESID_CHEM_CTRL_NML ${BLD}/CMAQ_Control_DESID_${MECH}_RED_EGU_POINT_NY.nml
Update the Spatial Masks for Emissions Scaling to use a file that contains state definitions for New York.
#> Spatial Masks For Emissions Scaling
setenv CMAQ_MASKS $SZpath/GRIDMASK_STATES_12US1.nc
Verify that the file contains New York
ncdump /shared/data/2018_12US1/surface/GRIDMASK_STATES_12US1.nc | grep NY
Output
float NY(TSTEP, LAY, ROW, COL) ;
NY:long_name = "NY " ;
NY:units = "fraction " ;
NY:var_desc = "NY fractional area per grid cell
Edit the output directory name in the#
# need to make the output directory prior to the beeond-cp
mkdir -p /shared/data/output/output_v54_cb6r5_ae7_aq_WR413_MYR_gcc_2018_12US1_DESID_REDUCE/LOGS
beeond-cp stagein -n ~/nodefile-$SLURM_JOB_ID -g /shared/data/output/output_v54_cb6r5_ae7_aq_WR413_MYR_gcc_2018_12US1_DESID_REDUCE -l /mnt/beeond/data/output/output_v54_cb6r5_ae7_aq_WR413_MYR_gcc_2018_12US1_DESID_REDUCE
Run CMAQ using DESID#
Note
The CMAQ run script has been configured to run on 192 cores (2 compute nodes of hb120v3 with 96 cores/node)
Change directories to the run script location
cd /shared/build/openmpi_gcc/CMAQ_v54/CCTM/scripts
Submit the Run script to the SLURM queue
sbatch run_cctm_2018_12US1_v54_cb6r5_ae6.20171222.2x96.ncclassic_DESID_RED_NY.csh
Check the status of the job
squeue
Output
1 hpc CMAQ lizadams CF 0:11 2 beeondtest2-hpc-[1-2]
Wait for the status to change from CF to R
Login to the compute node, install and run htop
ssh -Y IP-address
sudo yum install -y htop
htop
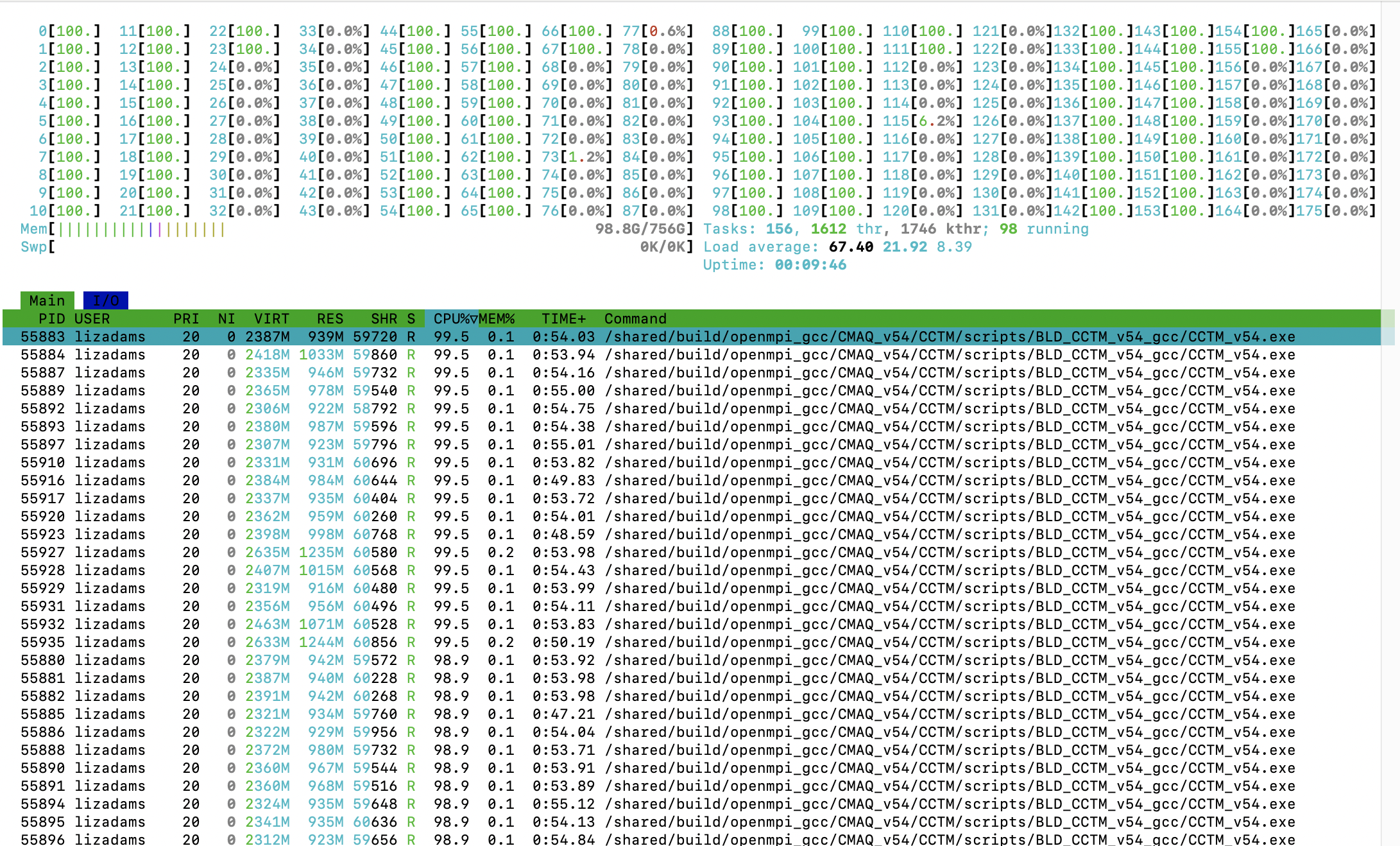
After the beeond copy copies the input data from /shared/data to the /mnt/beeond/data then CMAQ should start running. Htop should show that 96 processes are running and that 96G out of 756 G of memory is being used. ~
Review Log file from DESID run#
Note
The CMAQ run script has been configured to run on 192 cores (2 compute nodes of hb120v3 with 96 cores/node)
Review the Emissions Scaling Report Section in the CTM_LOG File to verify that for the NY region, the EGU emissions were scaled by 75%
cd /shared/build/openmpi_gcc/CMAQ_v54/CCTM/scripts
grep -A 20 'Stream Type: "Point Emissions File 2' CTM_LOG_001*
Output:
Stream Type: "Point Emissions File 2" | Sector Label: PT_EGU (04)
Table of Aerosol Size Distributions Available for Use Sector-Wide.
Note that Mode 1 is reserved for gas-phase species and emission variable.
Number Em. Var. Mode Reference Mode (see AERO_DATA.F)
------ -------------- --------------------------------
2 FINE FINE_REF
3 COARSE COARSE_REF
CMAQ Species Phase/Mode Em. Var. Region Op ScaleFac Basis FinalFac
------------ ---------- --------- ------ -- -------- ----- --------
NO2 GAS NO2 EVERYWHERE a 1.000 UNIT 1.000
NY o 0.750 UNIT 0.750
NO GAS NO EVERYWHERE a 1.000 UNIT 1.000
NY o 0.750 UNIT 0.750
HONO GAS HONO EVERYWHERE a 1.000 UNIT 1.000
NY o 0.750 UNIT 0.750
SO2 GAS SO2 EVERYWHERE a 1.000 UNIT 1.000
NY o 0.750 UNIT 0.750
SULF GAS SULF EVERYWHERE a 0.000 UNIT 0.000
NY o 0.750 UNIT 0.750